Full Text:
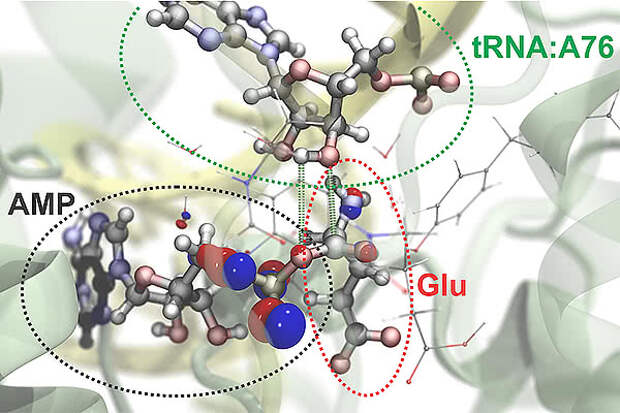
Scientists have built a "computational microscope" that can simulate the atomic and subatomic forces that drive molecular interactions. This tool will streamline efforts to understand the chemistry of life, model large molecular systems and develop new pharmaceutical and industrial agents, the researchers say.
The scientists combined two computational approaches used to simulate molecular interactions. The first, a nanoscale molecular-dynamics program known as NAMD, uses classical-mechanics methods to model the structure and simulate the behavior of hundreds of millions of individual atoms. The second program zooms in on the subatomic realm, simulating the interactions of protons, neutrons and electrons. Modeling at this quantum-mechanical scale demands a lot of computational power, so the researchers implemented a method for partitioning large molecules into classical- and quantum-mechanics regions. This allows them to focus their computational resources on small regions involved in critical interactions, such as the making or breaking of chemical bonds.Image credit: Rafael Bernardi, Zan Luthey-Schulten and Marcelo Melo
